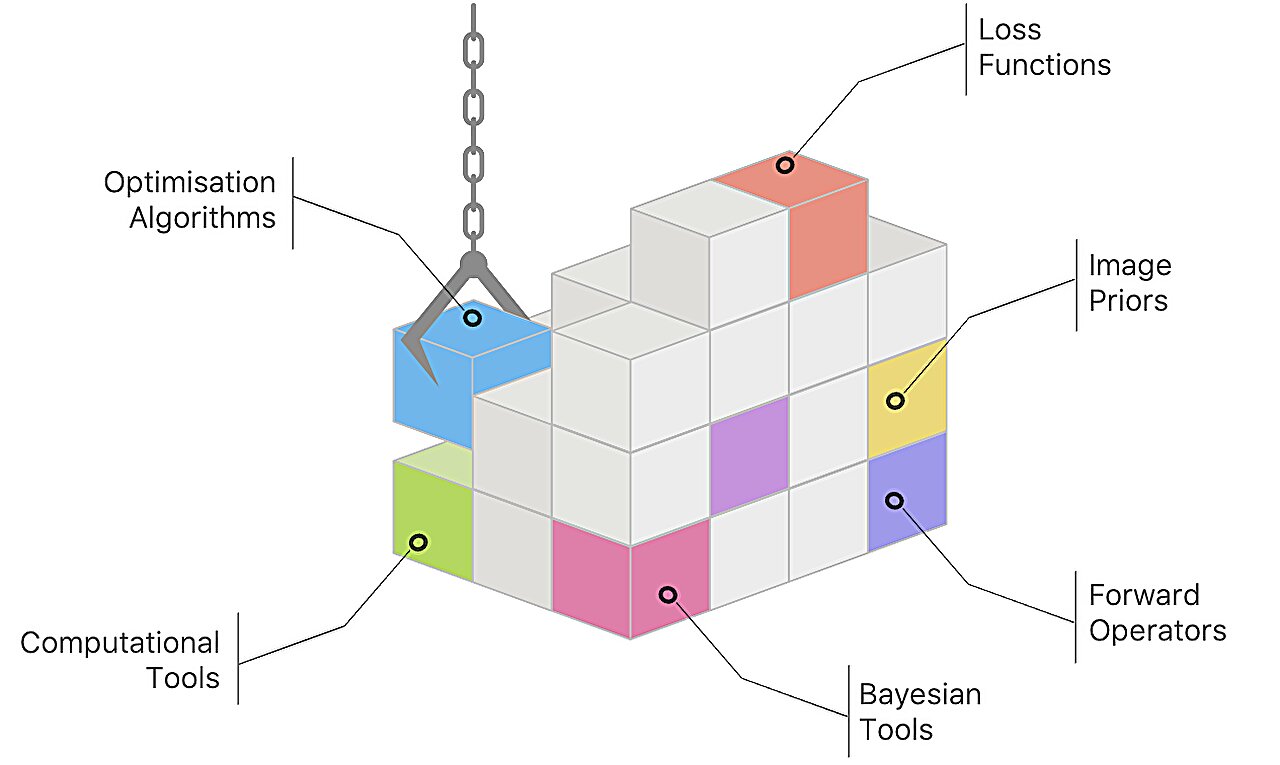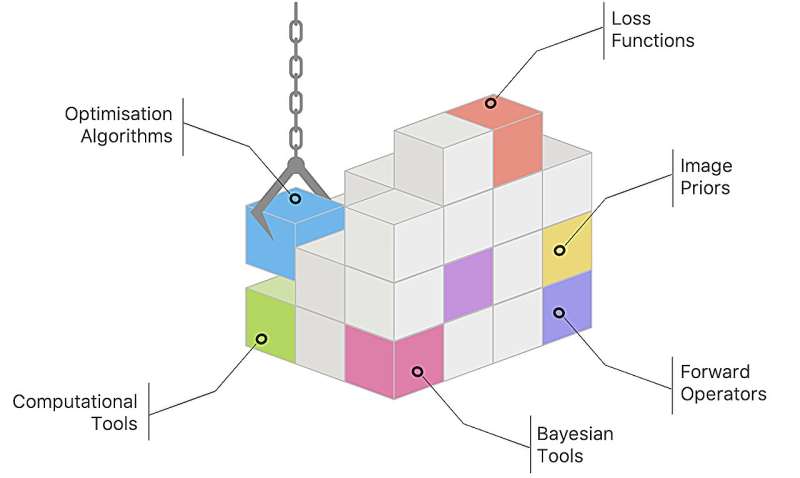
Scientists use an array of imaging instruments to look inside living organisms, sometimes as they move, and to observe inert objects without altering their state. Such instruments include telescopes, microscopes, CT scanners and more. But these instruments, even when working at maximum capacity, often generate only partial images or images of too low quality to provide much insight.
That's where powerful algorithms come in—they can piece together bits of missing information, improve an image's resolution and contrast, and flesh out sketchy objects. Impressive advances have been made recently in this technique, known as computational imaging, to the point where it now plays a central role in many types of research.
Engineers working in a variety of fields have developed powerful algorithmic programs for this technique, yet each one is designed for a highly specific application, even though the underlying imaging physics is generally the same. That means scientists wanting to combine imaging methods must make a considerable effort to adapt different programs and get them to communicate.
"We felt like we were always rewriting the same bits of code in order to adapt the programs we wanted to use," says Sepand Kashani, a Ph.D. student at EPFL's Audiovisual Communications Laboratory (LCAV).
So he teamed up with Matthieu Simeoni and Joan Rué Queralt, the former and current head of the Hub for Image Reconstruction at EPFL's Center for Imaging, to develop application-agnostic algorithms to be shared across different fields. Today that software, called Pyxu, is available in open source.
From tiny molecules to outer space, the same laws of physics apply
"The laws of physics governing imaging are often the same regardless of the particular field of research," says Rué Queralt. "And the problems encountered in image reconstruction can be grouped into a handful of categories with pretty much the same mathematical models—categories like X-rays and other forms of tomography, MRIs and radio astronomy, and so on." That's why he, Kashani and Simeoni believed it would be possible to develop application-agnostic software.
"Today, imaging methods are generally used only in the field they were initially developed for," says Rué Queralt. "We've seen scientists spend a lot of time and energy reinventing the wheel by coding programs similar to ones that already exist. That's slowing advancements in imaging across all areas."
Pyxu is intended to be used in any field and make it easier to seamlessly incorporate cutting-edge AI technology. Martin Vetterli, a professor at LCAV, explains: "Deep learning algorithms have upended the computational imaging landscape in recent years. These algorithms rely on AI technology and deliver better performance than their conventional counterparts."
The algorithms are trained by comparing high-quality images with reconstructed images, and then used to automatically make the corrections necessary to improve reconstructions and compare images themselves.
The Pyxu development team, consisting of engineers from both LCAV and the Center for Imaging, had to pool skills from a number of areas to create the software and open-source platform. "One of our biggest technical challenges was to make Pyxu flexible enough to process huge datasets yet easy to implement in a variety of IT systems with a broad range of hardware configurations," says Kashani.
Less code, more bricks
With Pyxu, scientists no longer need to be an expert in implementation details. The software contains modules representing different tasks, which users can select and piece together in the order they wish, much like Lego bricks.
Nino Hervé, a Ph.D. student at the University of Lausanne, was one of Pyxu's first users; he employed the software to reconstruct EEG images. "Interpreting the activity of 5,000 neural connections, based on readings taken by 200 electrodes placed on a patient's scalp, is no mean feat," he says.
"We need programs that are effective at solving optimization problems. Pyxu's software uses a variety of sophisticated optimization algorithms and is designed to run calculations in parallel, which makes it much faster. It's lightened my workload significantly."
Pyxu was released in open source just a few months ago and has already been used in numerous EPFL studies in fields such as radioastronomy, optics, tomography and CT scanning. "We designed Pyxu so that researchers could use our models as a basis for building their own," says Matthieu Simeoni, Pyxu's creator.
"Then the researchers can add their models to our software and make them available to the entire scientific community."
A second, more scalable version
A second, more scalable version of the software is currently in the works, with plans to release it too in open source. In addition to being able to handle larger datasets, the new version will include additional features and be even simpler to use. For instance, Pyxu's developers are working with engineers at EPFL's Biomedical Imaging Group to build on recent advances in embedding AI-driven algorithms into mathematical models.
The goal is to make sure reconstructed images convey important information visually and are mathematically robust—essential qualities for sensitive applications like medical diagnostics.
More information: Pyxu: pyxu-org.github.io/
Citation: Modular software for scientific image reconstruction (2024, May 2) retrieved 2 May 2024 from https://techxplore.com/news/2024-05-modular-software-scientific-image-reconstruction.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.