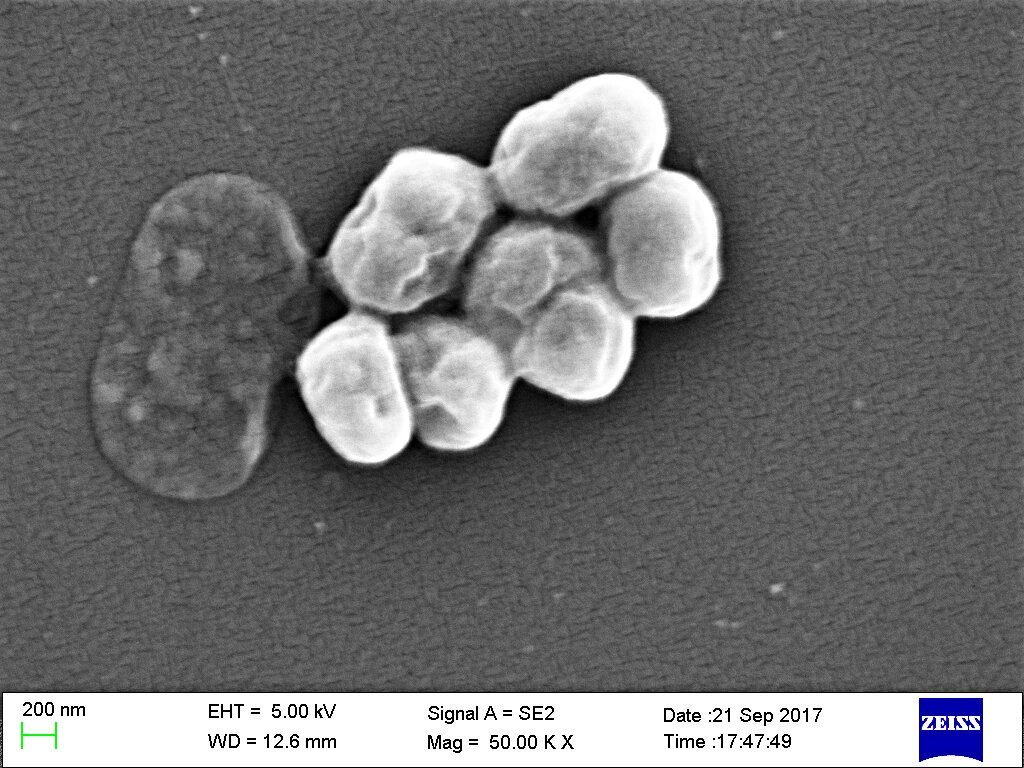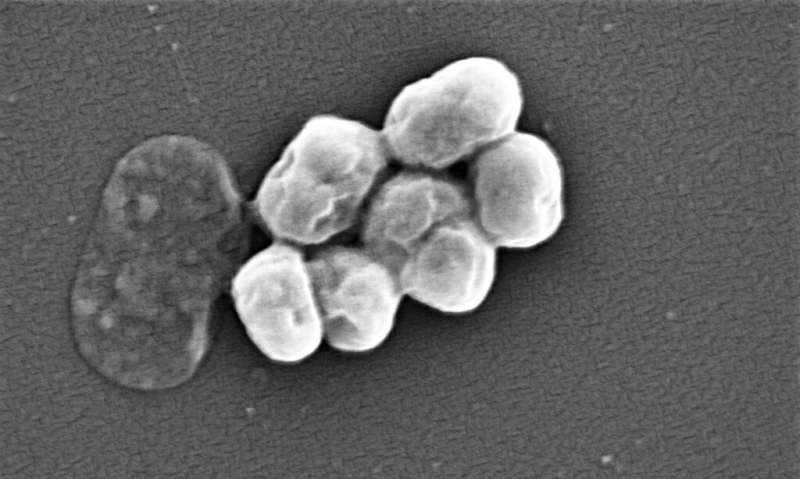
With nearly 5 million deaths linked to antibiotic resistance globally every year, new ways to combat resistant bacterial strains are urgently needed.
Researchers at Stanford Medicine and McMaster University are tackling this problem with generative artificial intelligence. A new model, dubbed SyntheMol (for synthesizing molecules), created structures and chemical recipes for six novel drugs aimed at killing resistant strains of Acinetobacter baumannii, one of the leading pathogens responsible for antibacterial resistance-related deaths.
The researchers described their model and experimental validation of these new compounds in a study published March 22 in the journal Nature Machine Intelligence.
"There's a huge public health need to develop new antibiotics quickly," said James Zou, Ph.D., an associate professor of biomedical data science and co-senior author on the study. "Our hypothesis was that there are a lot of potential molecules out there that could be effective drugs, but we haven't made or tested them yet. That's why we wanted to use AI to design entirely new molecules that have never been seen in nature."
Before the advent of generative AI, the same type of artificial intelligence technology that underlies large language models like ChatGPT, researchers had taken different computational approaches to antibiotic development. They used algorithms to scroll through existing drug libraries, identifying those compounds most likely to act against a given pathogen.
This technique, which sifted through 100 million known compounds, yielded results but just scratched the surface in finding all the chemical compounds that could have antibacterial properties.
"Chemical space is gigantic," said Kyle Swanson, a Stanford computational science doctoral student and co-lead author on the study. "People have estimated that there are close to 1060 possible drug-like molecules. So, 100 million is nowhere close to covering that entire space."
Hallucinating for drug development
Generative AI's tendency to "hallucinate," or make up responses out of whole cloth, could be a boon when it comes to drug discovery, but previous attempts to generate new drugs with this kind of AI resulted in compounds that would be impossible to make in the real world, Swanson said. The researchers needed to put guardrails around SyntheMol's activity—namely, to ensure that any molecules the model dreamed up could be synthesized in a lab.
"We've approached this problem by trying to bridge that gap between computational work and wet lab validation," Swanson said.
The model was trained to construct potential drugs using a library of more than 130,000 molecular building blocks and a set of validated chemical reactions. It generated not only the final compound but also the steps it took with those building blocks, giving the researchers a set of recipes to produce the drugs.
The researchers also trained the model on existing data of different chemicals' antibacterial activity against A. baumannii. With these guidelines and its building block starting set, SyntheMol generated around 25,000 possible antibiotics and the recipes to make them in less than nine hours. To prevent the bacteria from quickly developing resistance to the new compounds, researchers then filtered the generated compounds to only those that were dissimilar from existing compounds.
"Now we have not just entirely new molecules but also explicit instructions for how to make those molecules," Zou said.
A new chemical space
The researchers chose the 70 compounds with the highest potential to kill the bacterium and worked with the Ukrainian chemical company Enamine to synthesize them. The company was able to efficiently generate 58 of these compounds, six of which killed a resistant strain of A. baumannii when researchers tested them in the lab. These new compounds also showed antibacterial activity against other kinds of infectious bacteria prone to antibiotic resistance, including E. coli, Klebsiella pneumoniae and MRSA.
The scientists were able to further test two of the six compounds for toxicity in mice, as the other four didn't dissolve in water. The two they tested seemed safe; the next step is to test the drugs in mice infected with A. baumannii to see if they work in a living body, Zou said.
The six compounds are vastly different from each other and from existing antibiotics. The researchers don't know how their antibacterial properties work at the molecular level, but exploring those details could yield general principles relevant to other antibiotic development.
"This AI is really designing and teaching us about this entirely new part of the chemical space that humans just haven't explored before," Zou said.
Zou and Swanson are also refining SyntheMol and broadening its reach. They're collaborating with other research groups to use the model for drug discovery for heart disease and to create new fluorescent molecules for laboratory research.
More information: Kyle Swanson et al, Generative AI for designing and validating easily synthesizable and structurally novel antibiotics, Nature Machine Intelligence (2024). DOI: 10.1038/s42256-024-00809-7
Citation: Generative AI develops potential new drugs for antibiotic-resistant bacteria (2024, March 28) retrieved 28 March 2024 from https://techxplore.com/news/2024-03-generative-ai-potential-drugs-antibiotic.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.